Background:
Problem/Rationale:
Solution/Approach:
Table 1: Experimental design for the expression optimization
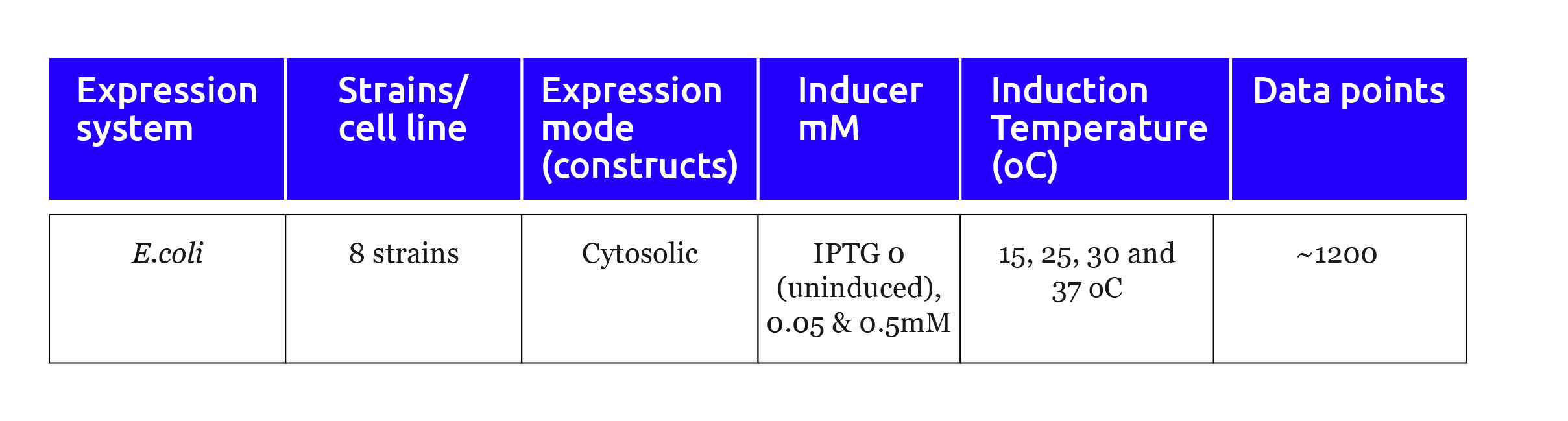
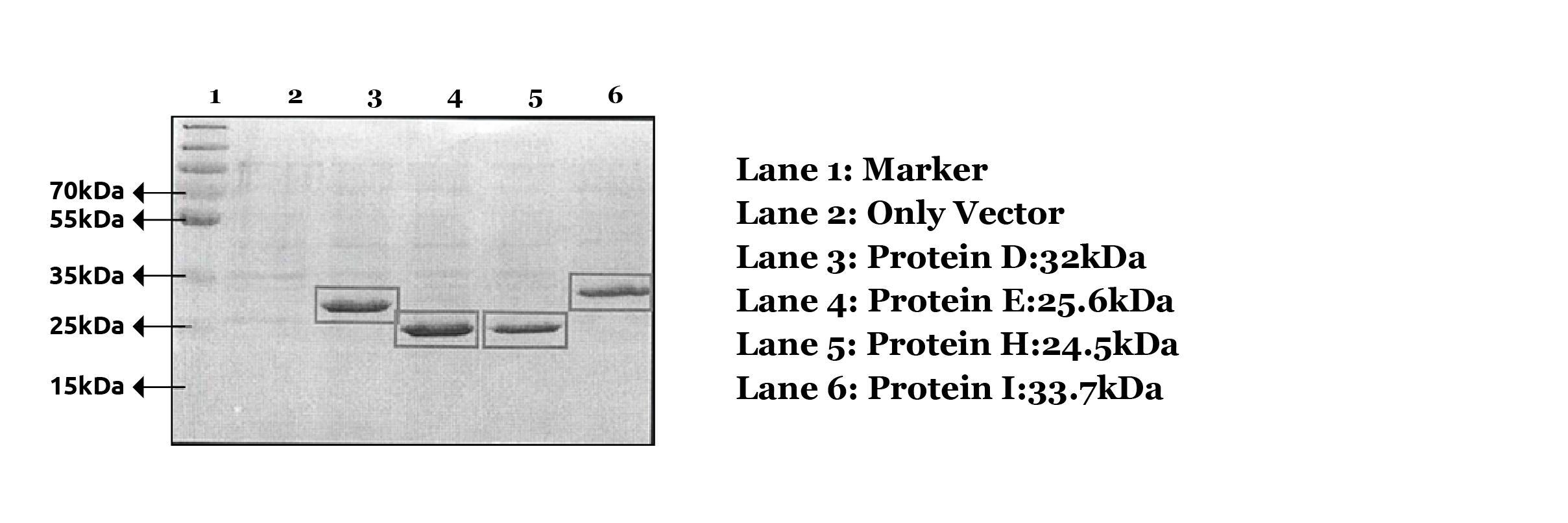
Figure 1: SDS-PAGE confirmation of four novel proteins
Measurable Impact:
Recommended reads
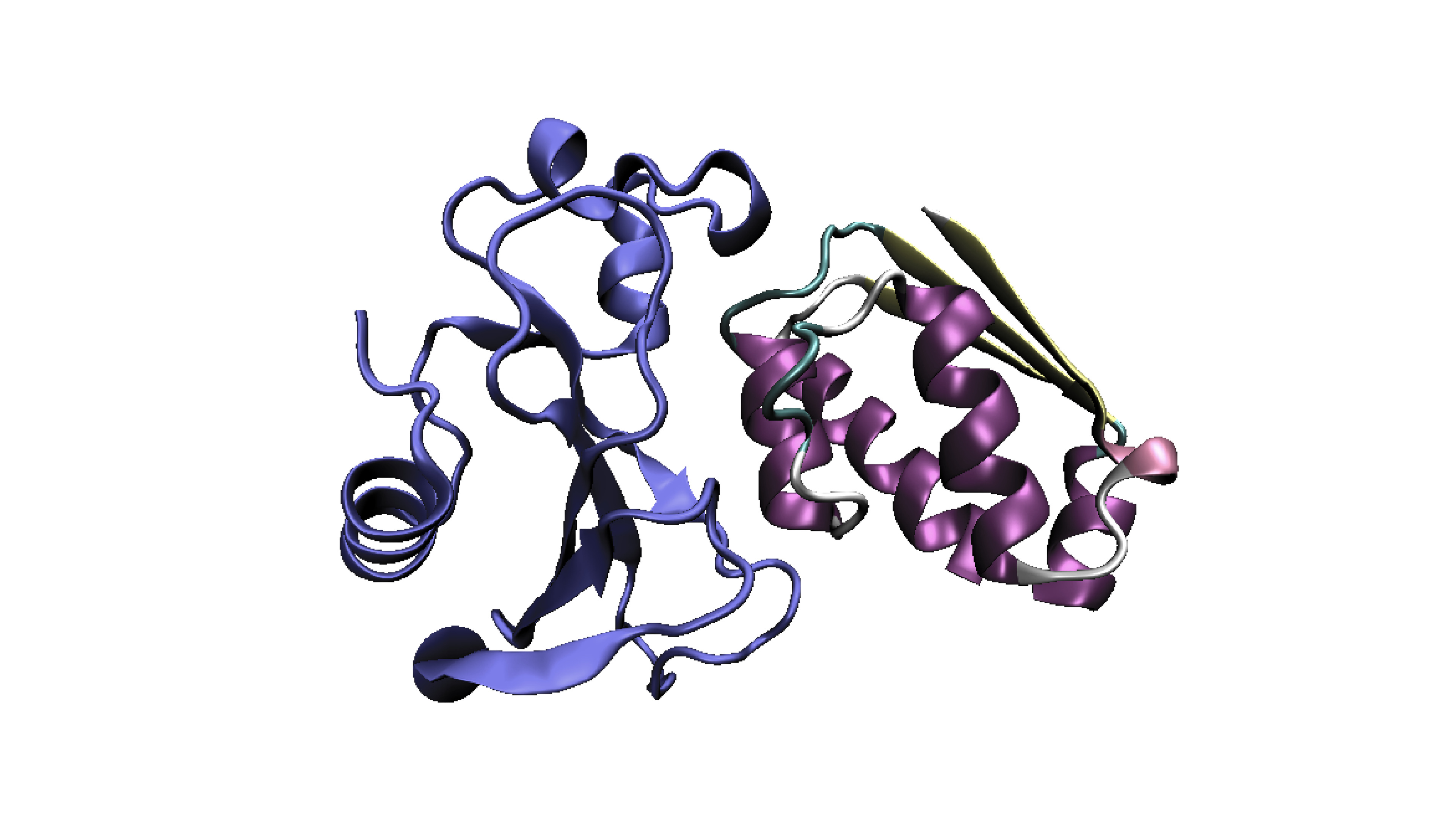
Expression and Purification of Highly Toxic Barnase Protein in Bacterial System
Magnitude of heterosis is much lower in self-pollinated crops than cross-pollinated crops. Therefore, improvement of self-pollinated crops by producing hybrid varieties are necessary and is a major goal of plant breeding. Several biotechnological strategies have been deployed to restrict self–fertilization in plants. However, the only commercialized transgenic male sterility method is SeedLinkTM,
Read More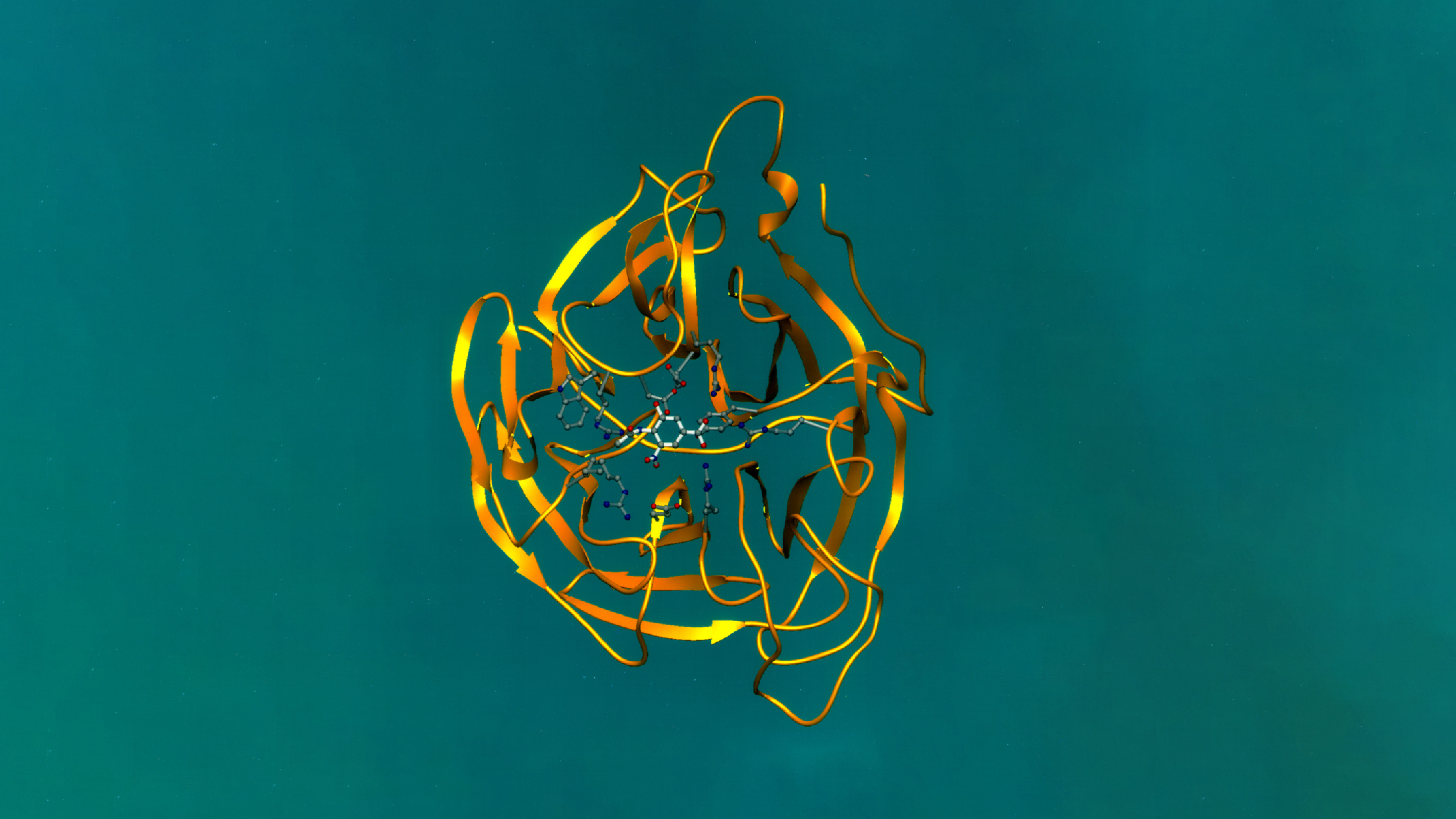
Expression of Full-Length Neuraminidase (NA)
Below is the case study describing the expression of full-length neuraminidase in the protease-deficient strain of Baker’s yeast, S. cerevisiae of D-Crypt™. The platform provides an ideal eukaryotic environment for expressing recombinant proteins retaining their correct structure, function, and post-translational modifications resembling those of humans. We used an array of proprietary expression vectors with designed upstream sequences for enhanced expression of neuraminidase.
Read More