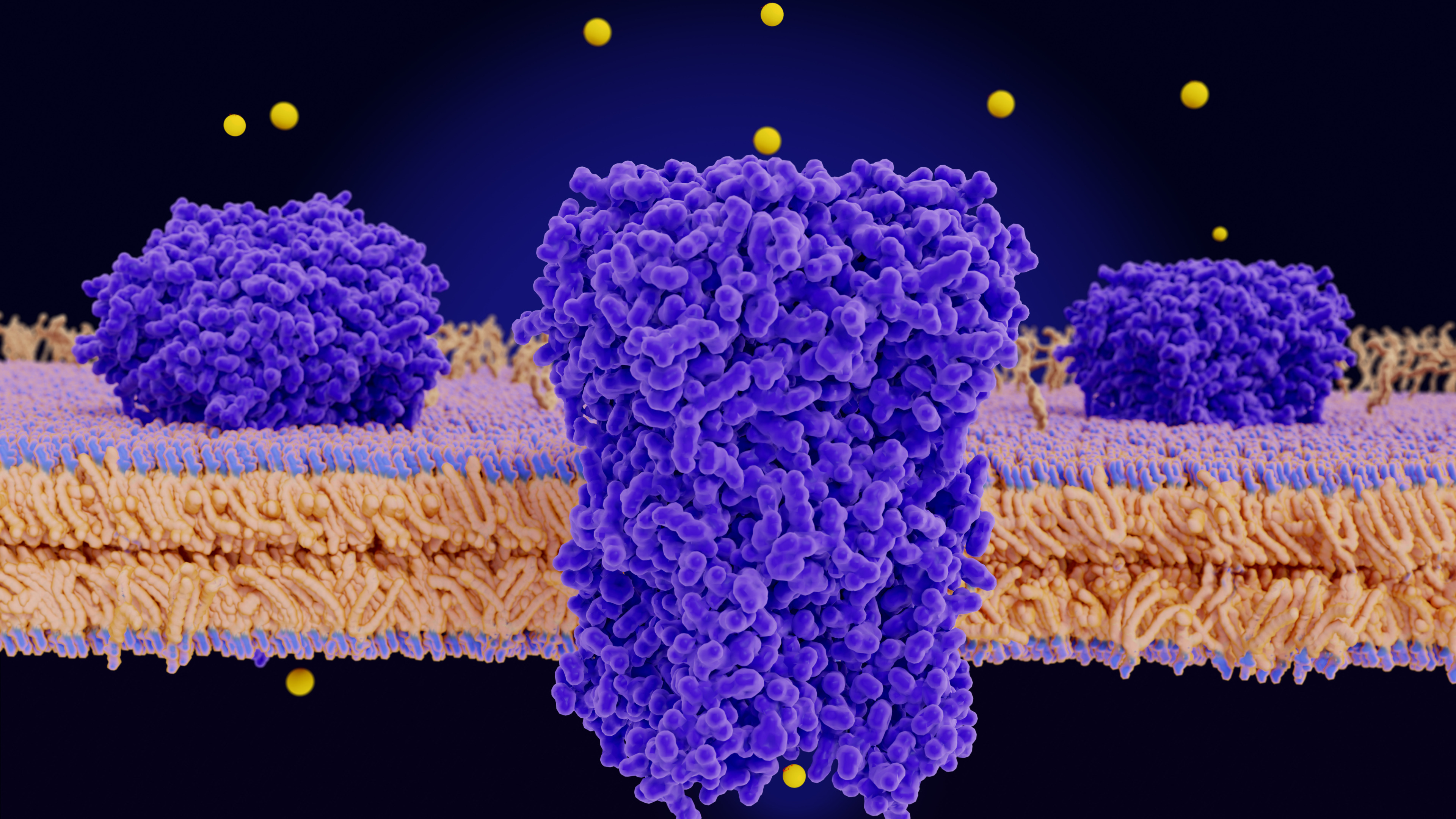
Background:
Problem/Rationale:
Solution/Approach:
Measurable Impact
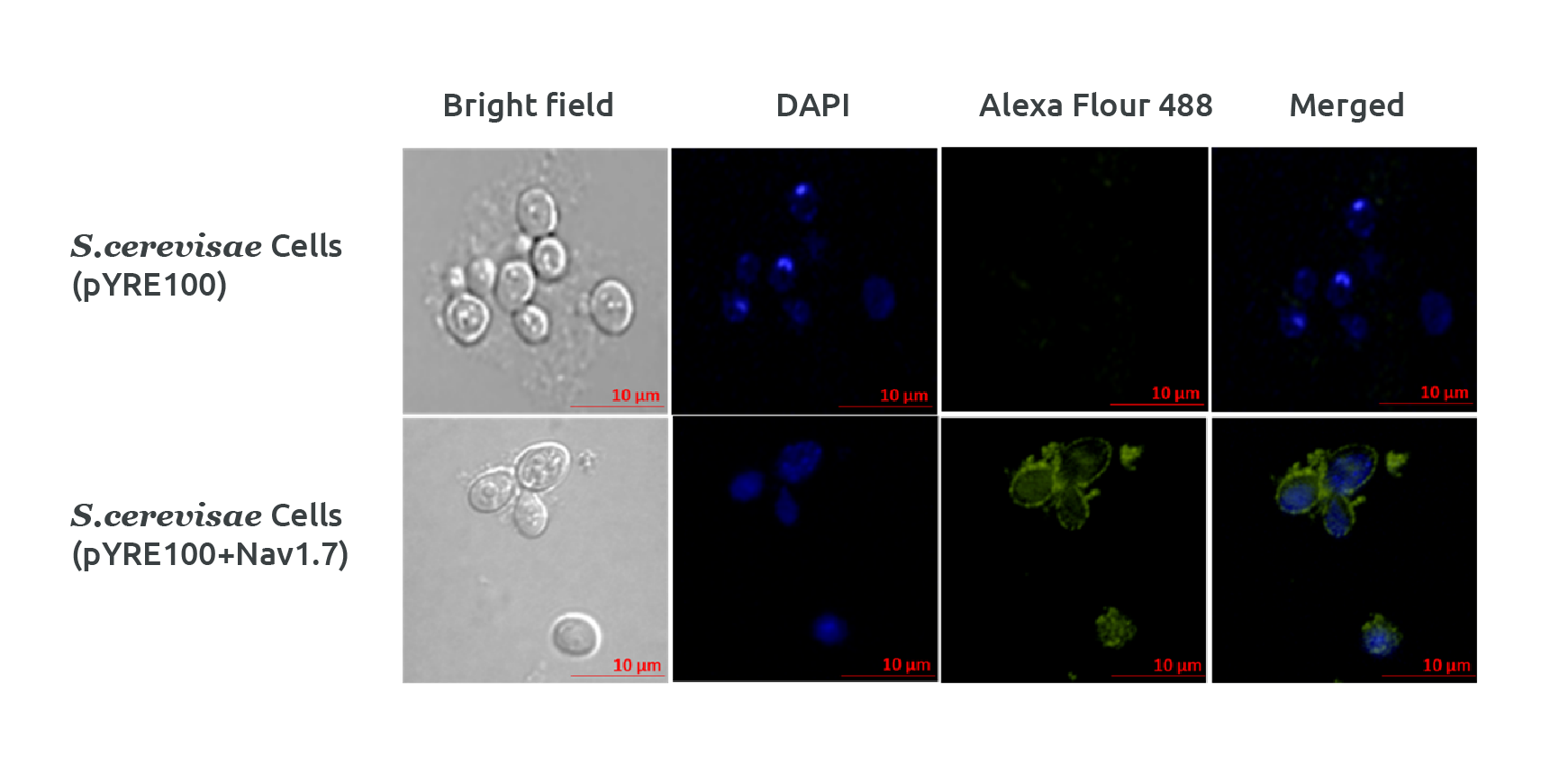
Figure 1: Confocal microscopy showing surface expression of NaV1.7
Recommended reads
Expression of Highly Active Protein in E. coli for Diagnostics Applicatio
The production of recombinant enzymes has led to the development of many diagnostics strategies in various medical fields. The increasing demand for recombinant proteins has created the need to develop efficient protein expression techniques to increase the productivity of the product.
Read More
Expression of Full-Length Neuraminidase (NA)
Below is the case study describing the expression of full-length neuraminidase in the protease-deficient strain of Baker’s yeast, S. cerevisiae of D-Crypt™. The platform provides an ideal eukaryotic environment for expressing recombinant proteins retaining their correct structure, function, and post-translational modifications resembling those of humans. We used an array of proprietary expression vectors with designed upstream sequences for enhanced expression of neuraminidase.
Read More
Purification Optimization and Large-Scale Production of a Recombinant Protein fr
The recombinant protein is a shortened form of the native isolate, requiring multiple tests and regulatory submission characterization. Some bacteria produce toxins that show specific activity against certain insect species. This specificity is mainly due to a unique binding site in the insects' midgut epithelium membrane, where the protein inserts and causes damage to the wall.
Read More